Roth Lab Research
Designing and engineering proteins
Chemogenetics:
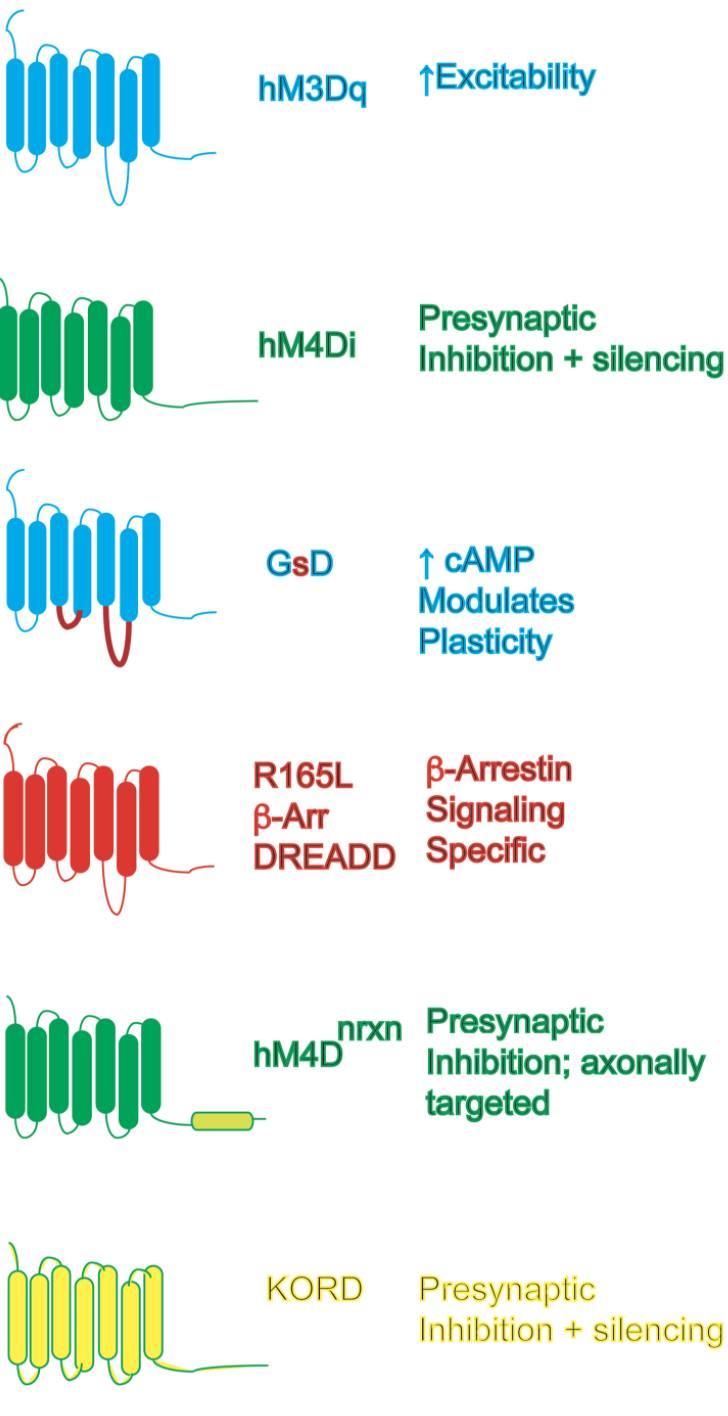
The Roth lab perfected the chemogenetic technology we have named “DREADD” (Designer Receptor Exclusively Activated by Designer Drugs; Armbruster et al, 2007). DREADD technology has afforded 1000’s of labs world-wide the opportunity to discover how cell-type specific modulation of signaling is translated into behavioral and non-behavioral outcomes (see Roth Neuron 2015 for recent review)
The Roth lab continues to enhance DREADD technology (see Vardy et al, Neuron 2015; Ngai et al, Nature Neurosci in press).
Directed Evolution:

We have created new technologies for directed molecular evolution in mammalian cells: VEGAS (Viral Evolution of Genetically Actuated Sequences; English et al, Cell 2019). This technology provides a platform for the evolution of mammalian proteins towards defined objectives and has been exemplified with transcription factors, nanobodies, and signaling molecules.
Nanobody Sensors:
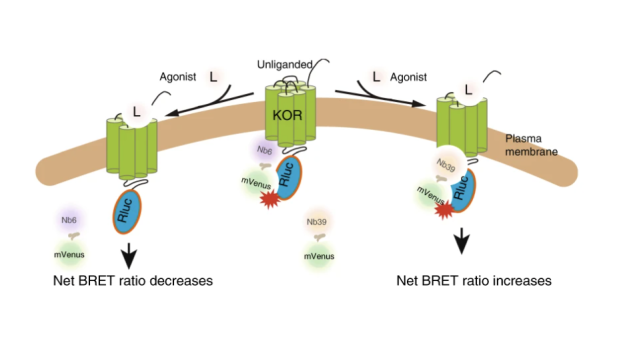
We have created nanobody based sensors to monitor the activation states of G protein coupled receptors (Che et al, Cell 2018; Che et al, Nature Comm 2020).
G Protein Sensors:
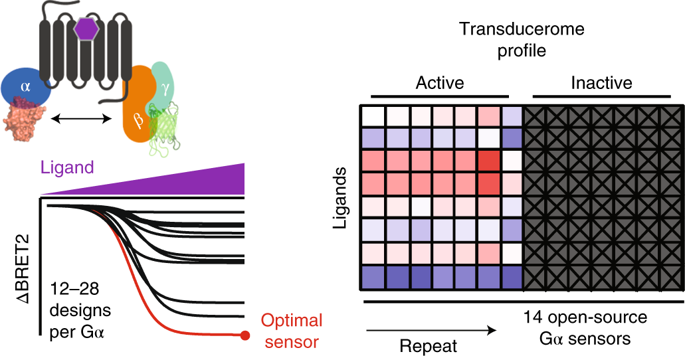
We have created a suite of BRET-based sensors to interrogate the transducerome for GPCRs (Olsen et al, Nature Chem Bio 2020)
Structural-guided Discovery of Novel Ligands for G Protein Coupled Receptors
Structural Elucidation of GPCR Structures:
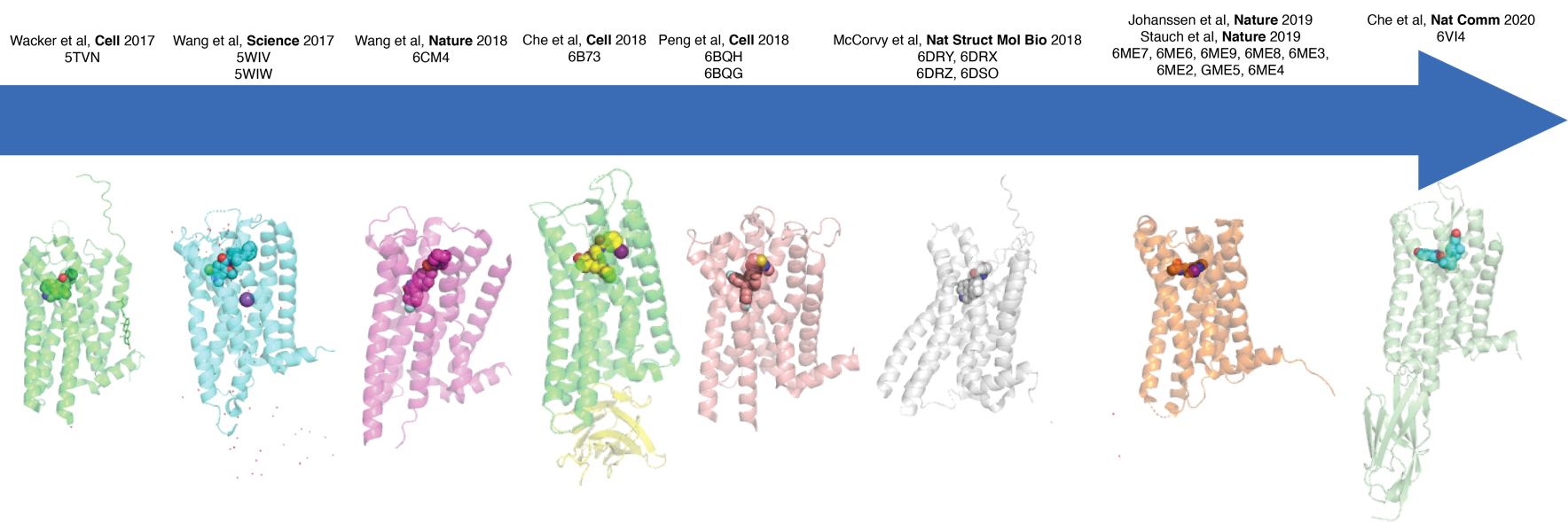
Recently solved GPCR structures include the structure of LSD bound to a serotonin receptor (Wacker et al, Cell 2018), the active (Che et al, Cell 2018), and inactive (Che et al, Nature Comm 2020) states of the kappa opioid receptors and many others (Figure below). We also have recently solved several structures of GPCRs in complex with transducers by cryo-EM.
Structure-guided Discovery of New Chemical Tools for GPCRs:
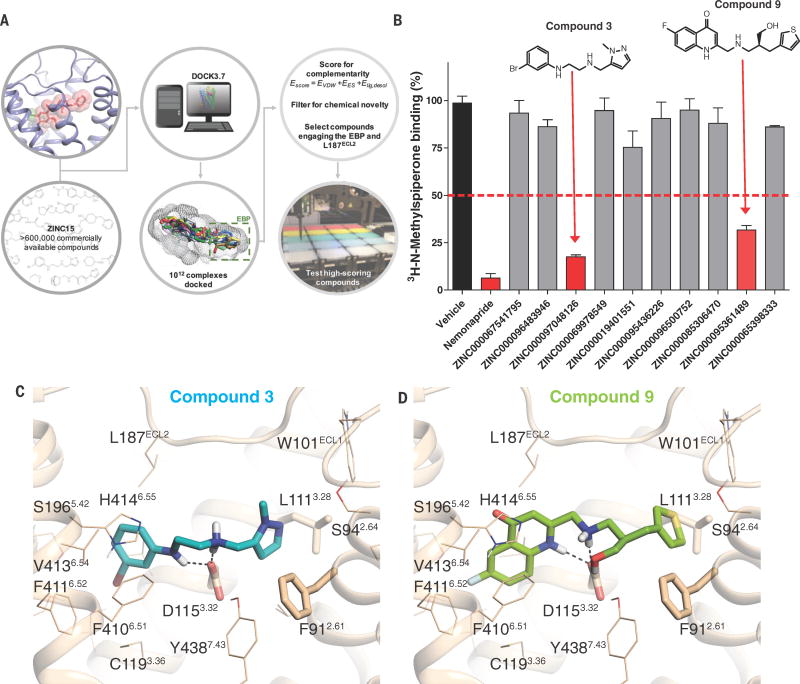
Using these high-resolution structures we have discovered novel chemotypes for a number of GPCRs including the D4-dopamine receptor (Wang et al, Science 2017; Lyu et al, Nature 2019), MT1 and MT2 melatonin receptors (Stein, Kang et al, Nature 2020) and D2 dopamine receptors (McCory et al Nature Chem Bio 2017).
Ongoing projects are aimed at utilizing these structures to discover new therapies for a number of neuropsychiatric disorders.